Hey,
The 9316600.sed file is a full SEED volume containing both headers and data records. The data records are what I call “bare” records, and technically not miniSEED because they do not include a Blockette 1000 that describes their length and data payload encoding. This was not uncommon with older data which were shipped as full SEED, where the SEED headers described the needed details.
To work data records without B1000 libmseed searches for signature of data records, thus skipping things it does not understand like SEED headers, and uses that search to determine record length. There is no way to determine the payload encoding, so libmseed simply has a default, which is Steim-1 because that is most prevalent with such older data. I believe the data payload decoding is failing either because 1) corruption, or much more likely, 2) the data are not Steim-1 encoded.
My normal, manual method of figuring out the encoding is to try them until I find one that decodes to something looking like a signal. In this case the SEED headers should contain the right details. Luckily rdseed still runs and can give a test output of a file like this:
% rdseed -s -f 9316600.sed:
B052F16 Format lookup: 1 Format Information Follows
B030F03 Format Name: REF32
B030F05 Data family: 0
B030F06 Number of Keys: 2
B030F07 Key 1: M0
B030F07 Key 2: W4 D0-23 C2
B052F17 Log2 of Data record length: 12
B052F18 Sample rate: 25
So the record lengths are 2^12 = 4096 bytes, check, and the data encoding is REF32. I’ve never heard of REF32, but the DDL keys, if you’re willing to squint at the SEED manual, describe non-multiplexed (M0), 24-bit integers (D0-23) in 32-bits of space (W4), using 2’s complement signing (C2).
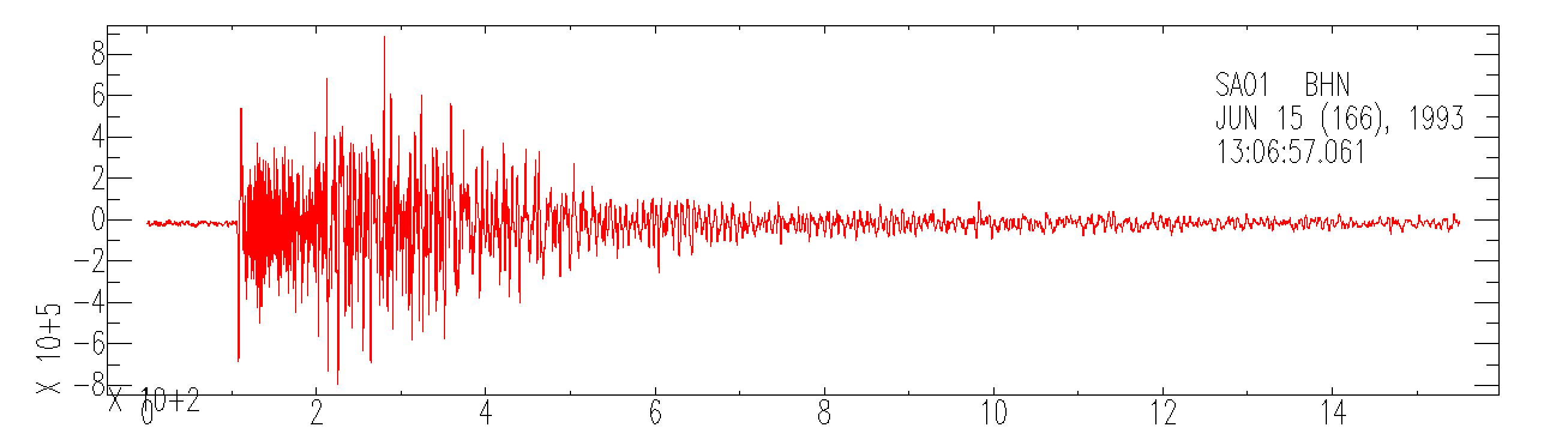
Based on this I tried decoding the data as 32-bit integers and got seismic-looking signals. For example with this command:
UNPACK_DATA_FORMAT=3 mseed2sac 9316600.sed
The data do not have network codes (introduced in the format in 1992) and so some software doesn’t handle that well. mseed2sac adds ‘XX’ as a network code to avoid having none.
Naturally I cannot guarantee the accuracy of the data interpreted as 32-bit integers and have only looked at a few samples.