Good afternoon, I’m Angel Matos from Peru, Here, this topic processing seismological data using Python and Obspy is not very widespread. I am currently working on my undergraduate thesis and a part of the workflow is to read a SEG2 file format recorded from an OYO seismograph (OYO Corp from Japan). I have two queries and I hope you can help me.
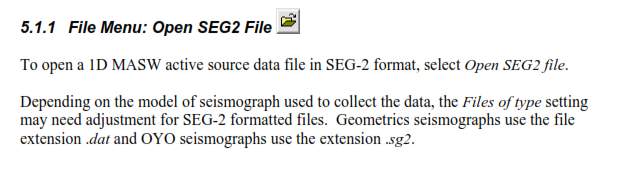
a) With Obspy, I can read files with .dat extension (from Geometrics seismograph), on the other hand, I can’t read files with .sg2 extension (from OYO seismograph). You can see from the picture below a better explanation about the extensions. This screenshot was taken from SeisImagerSW Manual
The case i that I don’t really want to use SeisImager, For my undergraduate thesis, I would like to read seismological data using Obspy, so, Is there a way I can read files with .sg2 extension.
Example of my code in Python (Python 3.8.5, Obspy 1.2.2, and working with JupyterNotebook on an environment created with Anaconda)
from obspy import read
st = read(pathname_or_url = “filename_1.dat”) #this works
st = read(pathname_or_url = “filename_2.sg2”) #this gives me a large error ending with KeyError: ‘SAMPLE_INTERVAL’
b) If that’s not possible, is there a way to convert .sg2 to .dat? Is it possible that exists a Python code for converting .sg2 to .dat? , since I need to automate the process (work with many files)
c) I would like you to be able to help me by providing a bibliography or some standard guide to how to work with these files. In a try to convert the files, I found this on internet:


I sincerely hope that you can help me because this is very important to me. Thanks in advance.