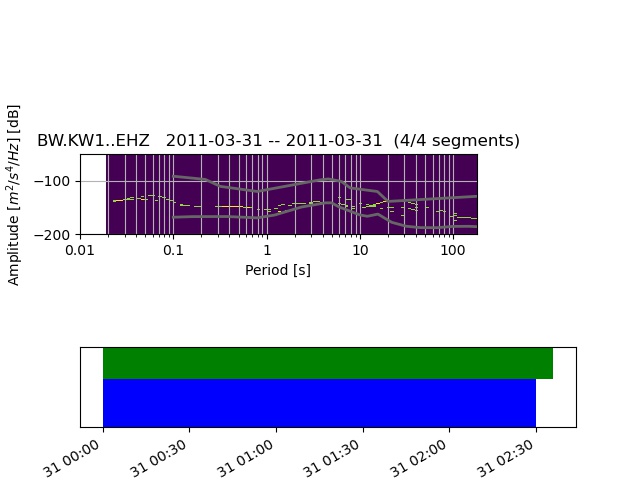
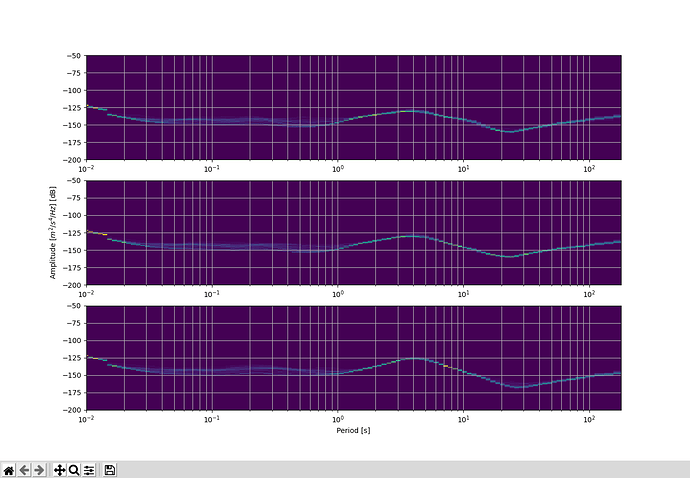
Trying to do a noise analysis for potential sites and want to create a single figure of 3 PPSD plots for each of the components stacked vertically. My code currently creates a individual PPSD figure. My approach originally was to create each figure and save those figures as a pickle file using
pklout = ppsd.plot(show=False) # this creates the figure and saves the output to pklout
pl.dump(pklout,open(tmppath + pklname,‘wb’)) # writes the Figure(640x480) to a pickle file
I wanted the then load each of the “pickled” figures back into memory in a different script to then stack each figure into a new figure vertically. Since the current PPSD function doesn’t appear to have the new high and low noise model as a plotting option (GitHub - ewolin/HighFreqNoiseMustang_paper), I also wanted to try and add these to the PPSD plots.
Can someone give me some tips on how to plot each of the individual component PPSD plots into a single plot and plot the new high/low noise models by Wolin and McNamara (2020)? I have the models from that paper as text files with 2 columns (period, dB level), so I figured that I could plot those with a simple matplotlib.pyplot “plot” command.
As an aside, are the figures generated with ppsd.plot() a similar object to that of a matplotlib plot? Is the figures I’m saving to pickle essentially just “raster” data like a jpeg or png such that I can’t modify those plots outside the ppsd.plot function?
Any help on this topic is greatly appreciated!
EDIT:
A thought I had might be to actually save the variable “ppsd” to a pickle file, then load that variable into my new script. Could I then do something to the effect of
from 1st script
ppsd = PPSD(tr.stats, metadata=inv)
ppsd.add(stm)
pickle.dump(ppsd,open(tmppath + pklname,‘wb’))
Start making figure
fig, axs = plt.subplots(3, 1)
for ii in range(3):
ppsd = pl.load(open(t_dflst.iloc[ii].item(), 'rb'))
axs[ii+1] = ppsd.plot(show_coverage=False, show=False) # mute show coverage because I want to keep figure uncluttered
plt.show()
After trying this last portion of code, what I found is that I create a blank figure with 3 subplots, but then each of the components ppsd’s are still plotted separately.